

Tapply, union, unique, unsplit, which.max, which.min Rank, rbind, Reduce, rownames, sapply, setdiff, sort, table, Mget, order, paste, pmax, pmax.int, pmin, pmin.int, Position, Grep, grepl, intersect, is.unsorted, lapply, Map, mapply, match,

The following objects are masked from ‘package:base’: anyDuplicated, append, as.ame, basename, cbind, colnames,ĭirname, do.call, duplicated, eval, evalq, Filter, Find, get, The following objects are masked from ‘package:stats’: IQR, mad, sd, var, xtabs Any advice for dealing with the genefilter issue? Thank you!! Please see error message below and session info.
SYMLINKER PERMISSION DENIED MAC MAC
I am on an MAC with M1 and am having similar issues. Old packages: 'RCurl', 'rmarkdown', 'xfun', 'XML'
SYMLINKER PERMISSION DENIED MAC INSTALL
Package which is only available in source form, and may need compilation of C/C++/Fortran: ‘DESeq2’ĭo you want to attempt to install these from sources? (Yes/no/cancel) no I've tried to force since it is installed already and then I tried none as you said. There are binary versions available but the sourceĭo you want to install from sources the package which needs compilation? (Yes/no/cancel)
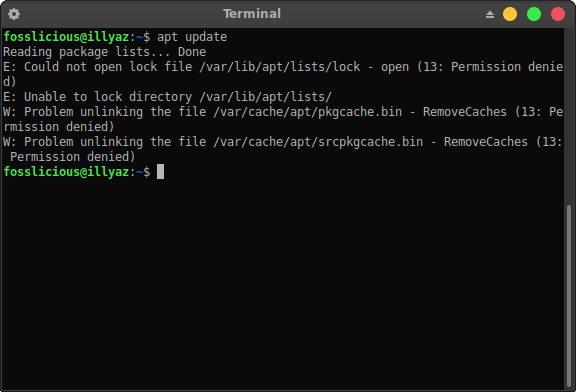
Warning: unable to access index for repository : 'getOption("repos")' replaces Bioconductor standard repositories,īioconductor version 3.13 (BiocManager 1.30.16), R 4.1.1Īlso installing the dependency ‘jquerylib’ Make: /opt/R/arm64/bin/gfortran: Permission denied Sudo ln -s /usr/local//gfortran/bin/gfortran /opt/R/arm64/binįinally BiocManager::install('genefilter') So i do this # make link to /opt/R/arm64/bin Still get make: /opt/R/arm64/bin/gfortran: No such file or directory Sys.setenv(gfortran="/usr/local/gfortran/bin") bash_profileĬOLLECT_LTO_WRAPPER=/usr/local/gfortran/libexec/gcc/x86_64-apple-darwin15/6.1.0/lto-wrapper The error showed: make: /opt/R/arm64/bin/gfortran: No such file or directory. Installation of package ‘genefilter’ had non-zero exit status
* removing ‘/Library/Frameworks/R.framework/Versions/4.1-arm64/Resources/library/genefilter’ Make: /opt/R/arm64/bin/gfortran: No such file or directoryĮRROR: compilation failed for package ‘genefilter’ opt/R/arm64/bin/gfortran -mtune=native -fno-optimize-sibling-calls -fPIC -Wall -g -O2 -c ttest.f -o ttest.o Ĭlang++ -arch arm64 -std=gnu++14 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c half_range_mode.cpp -o half_range_mode.oĬlang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c init.c -o init.oĬlang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c nd.c -o nd.oĬlang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c pAUC.c -o pAUC.oĬlang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c rowPAUCs.c -o rowPAUCs.oĬlang -arch arm64 -I"/Library/Frameworks/R.framework/Resources/include" -DNDEBUG -I/opt/R/arm64/include -fPIC -falign-functions=64 -Wall -g -O2 -c rowttests.c -o rowttests.o * installing *source* package ‘genefilter’. Installing the source package ‘genefilter’Ĭontent type 'application/x-gzip' length 805682 bytes (786 KB) Package which is only available in source form, and may need compilation ofĭo you want to attempt to install these from sources? (Yes/no/cancel) Yes 'getOption("repos")' replaces Bioconductor standard repositories, see '?repositories' for detailsīioconductor version 3.13 (BiocManager 1.30.16), R 4.1.0 () R studio BiocManager::install('genefilter') Macbook m1 R studio install deseq2 dependent package genefilter failed


 0 kommentar(er)
0 kommentar(er)
